From October 23-26, 2024, faculty and students from Utah’s BME department attended the annual Biomedical Engineering Society (BMES) national meeting in Baltimore, MD. Over 5500 registered attendees from 25 countries participated in a diverse technical, educational, industrial, and policy-related biomedical engineering program that featured topical, special, plenary and keynote sessions along with award forums and education and technology sessions in all-day parallel session formats. Industrial and government (e.g., NIH, NSF) speakers and panels also highlighted the annual event.
Four Utah BME faculty and 13 Utah BME undergraduate and graduate students attended to present their research and participate in networking opportunities with other participants in the BME community. Utah BME staffed an exhibit booth on the busy exhibition floor with over 100 other academic programs and relevant BME sponsors and partners.
Utah BME students presented the following posters or talks:
Utah undergraduate student poster presenters:
Chimdi V. Ihediwa
Co-authors: Clay T. Stanley (Electrical & Comp Eng PhD)
Faculty Advisor: Prof. Ashley N. Dalrymple
Poster Title: Image Processing of X-rays of the Lumbar Spine and Spinal Cord Stimulation Implants
Abstract: Neural injuries, such as spinal cord injury (SCI) and stroke, lead to significant motor impairments affecting individuals’ quality of life. Motor neuro-prostheses can vastly improve rehabilitation outcomes and increase independence and mobility. Spinal cord stimulation (SCS) has been FDA-approved for chronic pain management and was recently investigated in clinical trials for SCI rehabilitation, particularly for enabling locomotion. When SCS is used to treat pain, electrodes are typically placed over the dorsal columns, while for motor function recovery, electrodes are placed more laterally, targeting the dorsal roots.
The location of the electrodes is determined intraoperatively using fluoroscopy, with confirmation of electrode location relative to the vertebral segments in X-ray images. Of interest is pairing the anatomical electrode location with the functional output, measured as muscle activity using electromyography (EMG). However, current methods of manually identifying lead placement relative to the spinal vertebral level via X-ray images are time-consuming and prone to inaccuracies. This compromises the ability to accurately correlate muscle activity from EMG with the location of the corresponding SCS electrode. To overcome this challenge, we propose to use image analysis and machine learning methods to automate the detection of electrode location relative to the spinal vertebral level. Additionally, we will test a new electrode approach in cadavers to better target motor function. Automated image processing will enable us to precisely correlate evoked motor responses with specific spinal cord levels where the stimulation was applied, thus enhancing the effectiveness of SCS for targeted muscle activation and improving rehabilitation outcomes.
Daniel Candland
Co-authors: Mahima Choudhury, PhD, BME and James Craig (BME Lab technician)
Faculty Advisor: Prof. Tara Deans
Poster title: Determining Transcription Factor Dynamics During Osteogenesis
Abstract: The ability of Mesenchymal Stem Cells (MSCs) to self-renew and differentiate into many cell types makes them a prime candidate for use in regenerative medicine, however the ability to direct stem cell fate for tissue-specific applications remains challenging. Stem cell fate is controlled by a complex network of Gene Regulatory Networks that respond to stimuli through dynamic changes in expressed proteins that can turn genes on or off, called transcription factors. Synthetic biology offers a method of controlling stem cell fate through reprogramming cells with genetic circuits that can mimic the dynamic effects of gene regulatory networks on command. Osteogenesis, the process in which MSCs differentiate into bone tissue, offers a potential opportunity to examine, decipher, and utilize the dynamic transcription networks behind differentiation. A major hurdle in understanding osteogenic differentiation is the dynamic nature of these transcription factor patterns, which consist of multiple genes interacting spatially and temporally during the process. Recent advancements in machine learning, a kind of Artificial Intelligence (AI) that can estimate statistical patterns based on input data, can be used to overcome this hurdle. By first building a library of expression data for key genes of interest across multiple time points during osteogenesis, machine learning can be used to analyze the data and form an understanding of how transcription factor expression is regulated in cells during differentiation. This can be used to build genetic tools that mimic these dynamic expression patterns to reprogram cells and gain control over osteogenic differentiation.
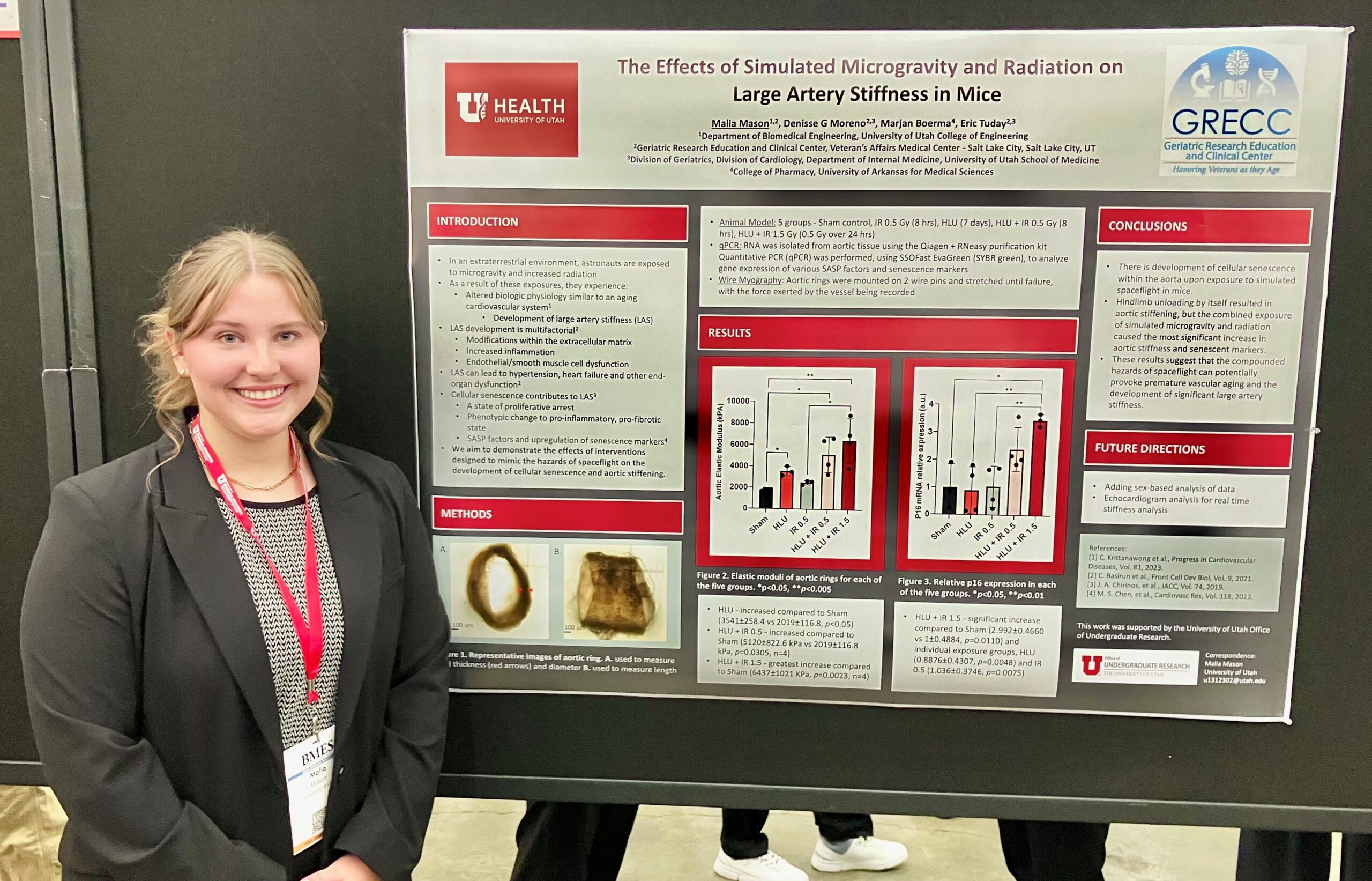
Malia Mason
Co-authors: Denisse G Moreno and Marjan Boerma
Faculty Advisor: Eric Tuday
Title: The Effects of Simulated Microgravity and Radiation on Large Artery Stiffness in Mice
Abstract: Astronauts often return from the extraterrestrial environment, after experiencing atypical exposures such as microgravity and increased radiation, with altered biologic physiology. Observations have been made that some of these physiologic changes bear some similarities to that of the aging of the cardiovascular system, in particular the development of large artery stiffness (LAS). The development of LAS is multifactorial and includes modifications within the extracellular matrix, increased inflammation, and endothelial/smooth muscle cell dysfunction that can lead to hypertension, heart failure, and other end-organ dysfunction. Cellular senescence is characterized as a state of proliferative arrest, and a change of the cellular phenotype to a non-functional, pro-inflammatory, and pro-fibrotic state which has been shown to contribute to the development of LAS. Recent evidence suggests that exposure to the conditions of spaceflight, microgravity, and galactic cosmic radiation have the potential to induce cellular senescence and lead to premature vascular aging and LAS. Therefore, we aim to demonstrate the effects of interventions designed to mimic the hazards of spaceflight on the development of cellular senescence and examine their correlation to aortic stiffness
Utah BME graduate student presenters
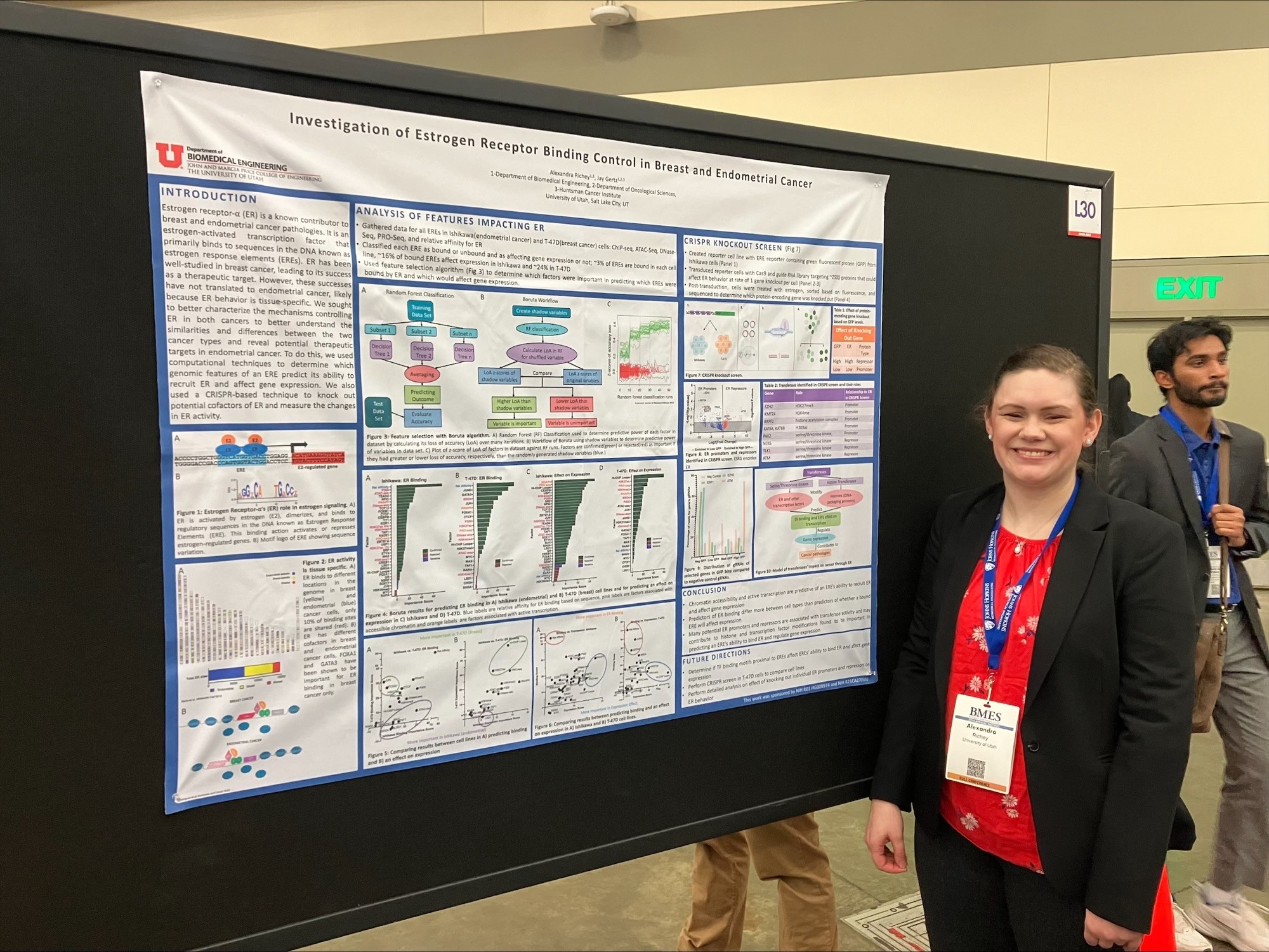
Alexandra Richey
Faculty Advisor: Prof. Jay Gertz
Title: Investigation of Estrogen Receptor Binding Control in Breast and Endometrial Cancer
Abstract: Endometrial cancer is the most common gynecologic cancer in the United States and the fourth most common cancer in women. Incidence and mortality are both rising, and five-year survival rates have decreased since the 1970s. Most endometrial cancers are estrogen receptor alpha (ER) positive and are related to excess estrogen signaling. ER is an estrogen-activated transcription factor that mostly binds to sequences in the DNA known as estrogen response elements (ERE) to regulate gene expression. It is also a known contributor to many breast cancer pathologies, and many therapeutics targeting ER in breast cancer have been successfully developed. However, these successes in breast cancer have not translated to endometrial cancer, likely because ER exhibits tissue-specific behavior. ER has been shown to bind to different genomic locations and to have different cofactors in breast and endometrial cancer cells. However, much is still unknown about the extent to which ER behavior differs between the tissues. We seek to better characterize the mechanisms controlling ER behavior in both breast and endometrial cancer cells. To do this, we will use computational techniques to determine which genomic features of an ERE predict its ability to recruit ER and affect gene expression. We will also use a CRISPR-based experimental technique to knock out potential cofactors of ER and measure the changes in ER binding and gene expression. This will allow us to better understand the mechanisms of ER binding control, and how they differ between the cell types.
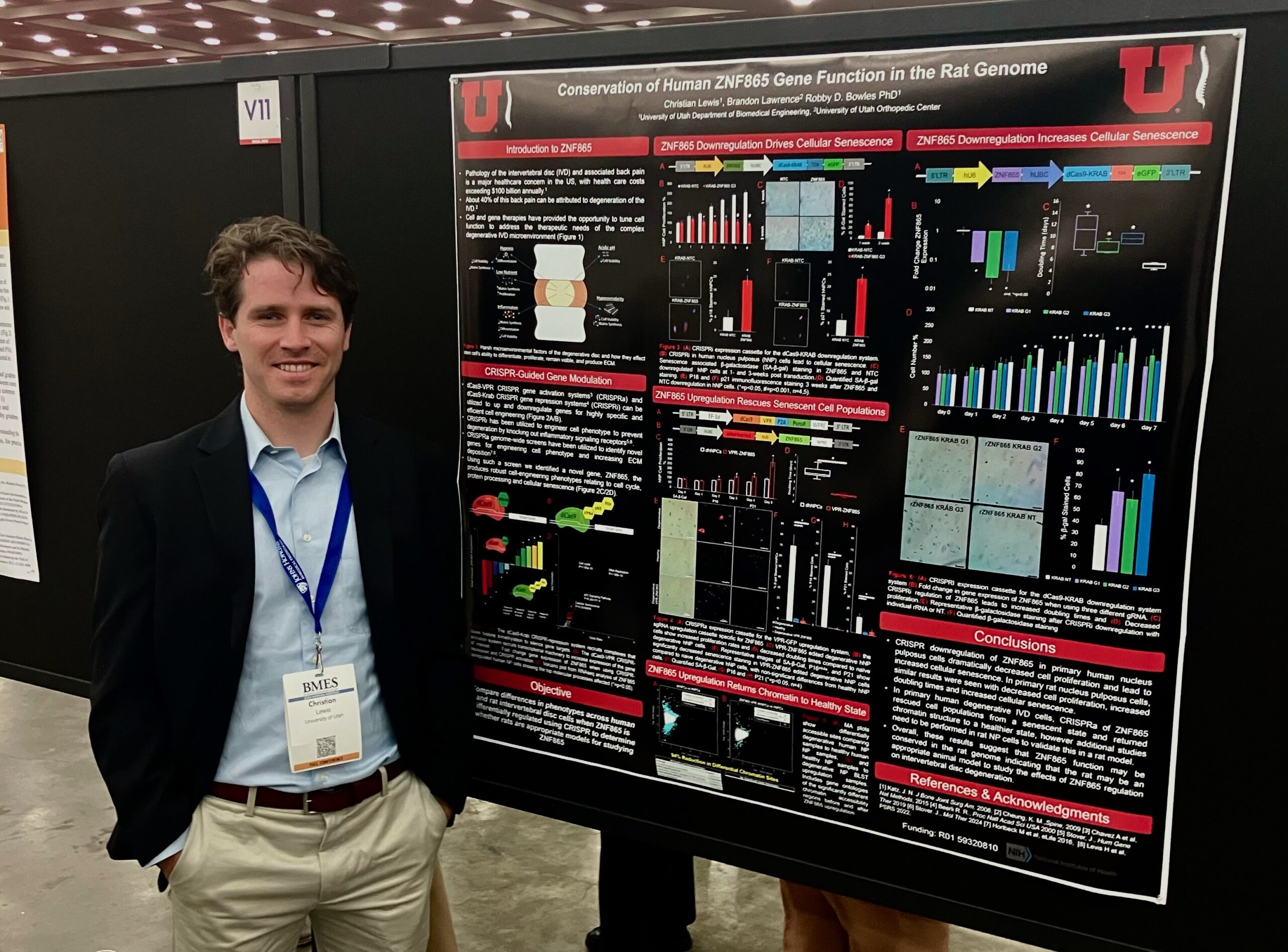
Christian Lewis
Co-authors: Jake Polaski, (BME Postdoc Research Associate),
Faculty Advisor: Profs. Brandon Lawrence and Robby D. Bowles.
Title: CRISPR Regulation of ZNF865 Rescues Human Nucleus Pulposus Cell Populations from Senescence
Abstract: Pathology of the intervertebral disc (IVD) and associated back pain is a major healthcare concern in the US with health care costs exceeding $100 billion annually. About 40% of this back pain can be attributed to degeneration of the IVD. Degeneration of the IVD has been closely associated with IVD cell senescence, with many cells present in degenerative IVD tissue demonstrating a senescent state. Current treatments for IVD degeneration are mainly palliative, aimed at reducing pain, providing a critical need for the development of novel treatments for IVD degeneration. Cell engineering has provided the opportunity to tune cell function to address therapeutic needs and discover novel biology. Recently, in a set of CRISPRa genome wide screens, our lab identified a novel zinc finger protein previously unpublished on, ZNF865, referred to here as BLST, that produces robust cell-engineering phenotypes relating to cell cycle, protein processing and cellular senescence. Preliminary data from publicly available RNAseq datasets suggested that BLST is differentially regulated within degenerating human IVDs. Here we investigate BLST within primary human IVD samples to determine relative levels of gene expression within degenerative IVDs and examine the ability for CRISPR regulation of BLST to regulate senescence within these human primary IVD cells.
Diego Perez
Faculty Advisor: Prof. A. Chuck Dorval
Title: Temporal Characteristics of Speech in Cortical and Subcortical Recordings
Abstract: Speech requires complex motor planning for activating coordinated jaw, tongue, lips, and larynx muscles. Motor-associated areas of the cortex, particularly frontal regions, are known to drive speech motor planning and execution. However, little is known about subcortical regions’ roles in speech production. The basal ganglia is one subcortical region engaged in motor planning. Disorders of the basal ganglia, such as Parkinson’s disease (PD), can lead to significant speech production-related issues (e.g., dysarthria, monotone). Furthermore, electrophysiological recordings of the subthalamic nucleus — a subregion within the basal ganglia and a common stimulation target for the surgical management of PD — have shown a decrease in 15–30 Hz “beta” power (Hebb et al., 2012) and an increase in 70–150 Hz “high-gamma” power (Chrabaszcz et al., 2019) during speech production.
Nevertheless, the relationship between the cortical and subcortical regions in speech production remains poorly understood. In this study, we utilize simultaneous local field potential recordings from the cortical and subcortical regions of volunteers undergoing awake Deep Brain Stimulation (DBS) surgery for PD and Essential Tremor (ET) during a three-speech-syllable repetition task. We investigated possible communication mechanisms by calculating synchronization between the cortical and subcortical regions and identifying the direction of any information transfer during speech production.
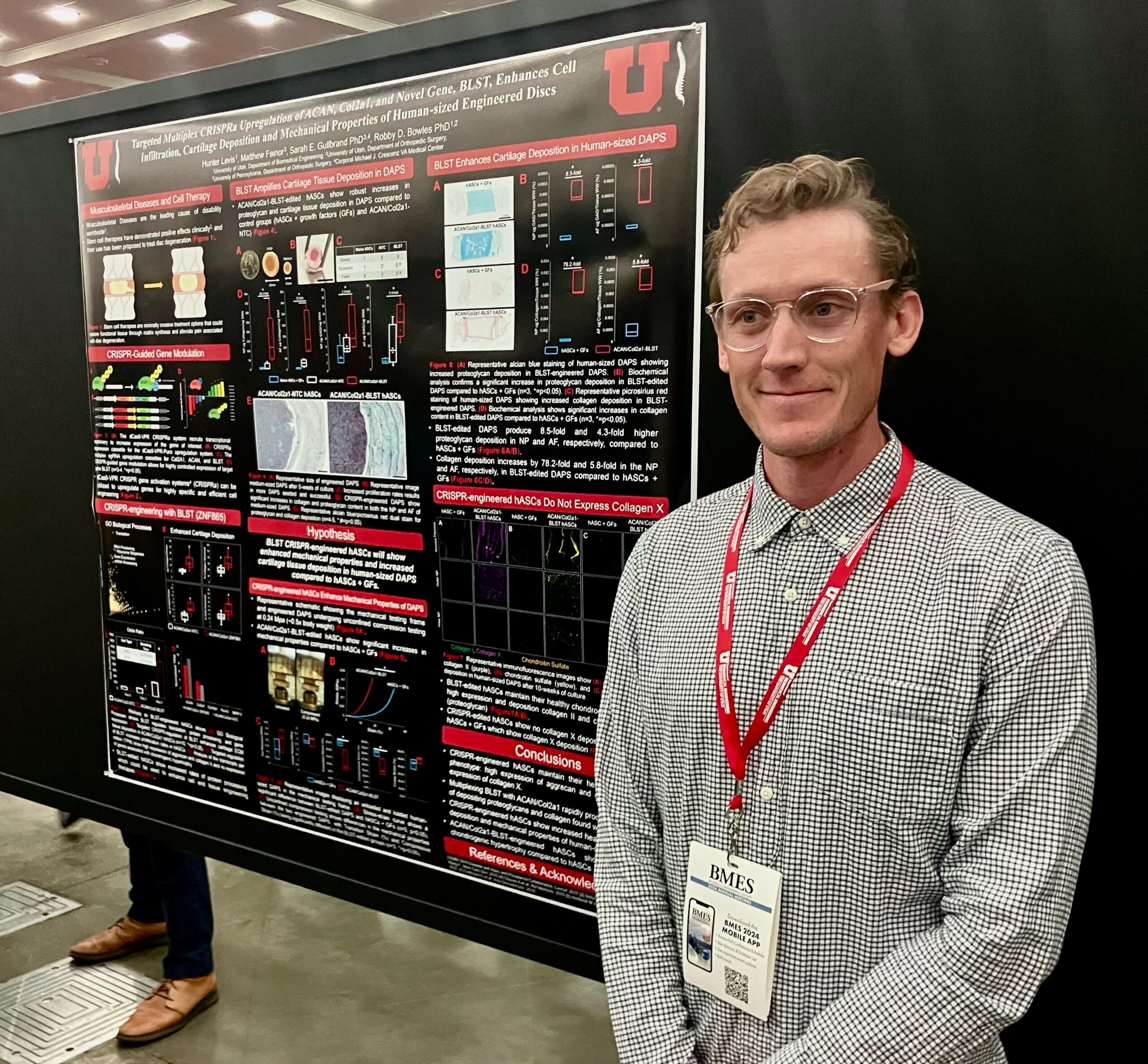
Hunter Levis
Co-Authors: Matthew Fainor, Ameerah Lawal, and Sarah E. Gullbrand
Faculty Advisor:Prof. Robby D. Bowles
Title: Targeted Multiplex CRISPRa Upregulation of ACAN, Col2a1, and Novel Gene, ZNF865, Enhances Cell Infiltration, Cartilage Deposition and Mechanical Properties of Human-sized Engineered Discs
Abstract: Recently, a CRISPR-activation (CRISPRa) screen led to our lab’s discovery of a previously unknown zinc finger protein, ZNF865. ZNF865 regulates cell senescence, activity, and protein processing which has shown potential as a cell engineering tool. Disc-like angle ply structures (DAPS) are artificial total intervertebral disc (IVD) replacements that have shown promise as therapeutic interventions to treat severe cases of degenerative disc disease (DDD). Here we are testing ZNF865’s ability to amplify cell phenotype using CRISPRa. Utilizing targeted CRISPRa upregulation of aggrecan (ACAN), collagen-II (Col2a1), and ZNF865 in human adipose-derived stem cells (hASCs), we hypothesize that we can improve cartilaginous ECM deposition and the mechanical properties of large human-sized DAPS.
Jérémi Godbout
Faculty Advisor: Prof. A. Chuck Dorval
Title: Temporal Interference Simulation Drives Polarization in a Computational Neuron Model
Abstract: Temporal interference stimulation (TIS) was developed as an alternative method of transcranial brain stimulation to noninvasively modulate neural circuits deep in the human brain. Existing clinical transcranial brain stimulation — such as transcranial alternating current (tACS), direct current (tDCS), and magnetic (tMS) stimulation — share a common fundamental limitation: a trade-off between brain depth, neural target focality, and activating field intensity. The TIS approach has the potential to circumvent this trade-off, enabling deep and focal neural activation. TIS leverages two “carrier” frequencies offset by a small frequency difference that produced a neuromodulatory “beat” pattern at the difference frequence. This beat field can peak deep in the brain, can be more focal than conventional stimulation, and can be steered into specific regions by adjusting the relative intensities of the carrier fields. Modeling such stimulation onto biophysically realistic single cell models is the first step to understanding the effects, possible applications, and potential limitations of TIS.
Madison Lodico
Faculty Advisor: Prof. A. Chuck Dorval
Title: Axon Geometry Impacts Activation Threshold: Implications for Deep Brain Stimulation\
Abstract: Parkinson’s disease (PD) is the second most common neurodegenerative disorder in the United States and the number of Americans with PD is expected to double by 2040 according to the National Institute of Neurological Disorders. Deep brain stimulation (DBS) is a neurosurgical procedure used to treat many movement disorders such as Parkinson’s disease as well as other neurological conditions such as essential tremor, epilepsy, and dystonia. However, the success of therapeutic electrical stimulation varies greatly between patients and across neurological disorders. According to Chaturvedi et al, a primary factor for this is electrode misplacement, one of the biggest sources of error in DBS treatment. One unaccounted-for factor that could significantly contribute to inaccurate electrode placement stems from using the universally accepted straight axon model to estimate neuronal excitability from as a function of contact location. Past work, such as Abdeen et al, relating to the success of DBS have typically assumed that using geometrically straight versus biologically meandering axon models is inconsequential. In this work, we quantify the changes in neuronal excitability as a function of axon curvature, independent of all other variables.
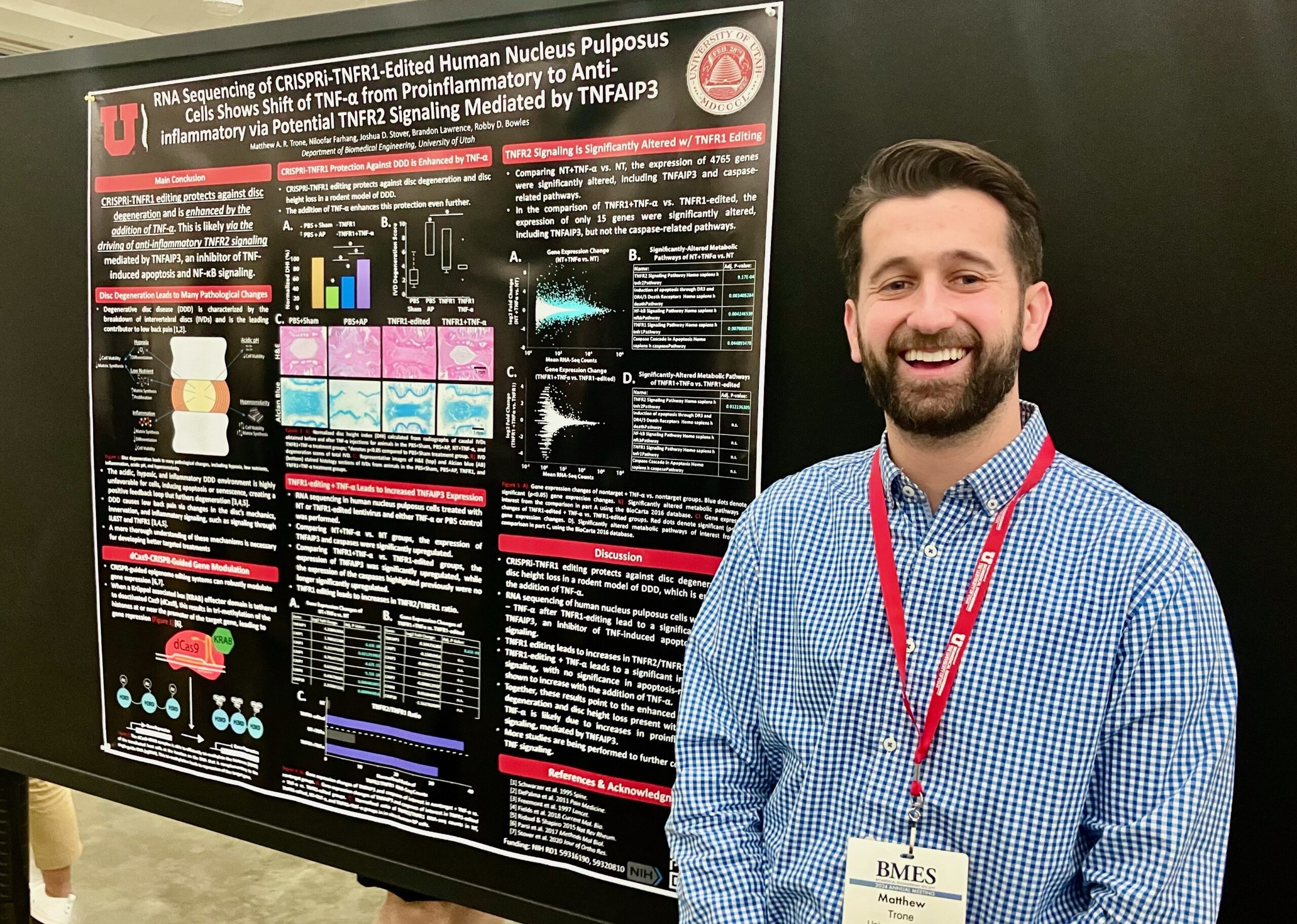
Matthew A. R. Trone
Co-authors: Niloofar Farhang, PhD, BME and Joshua D. Stover, PhD, BME
Faculty Advisor: Profs. Brandon Lawrence and Robby D. Bowles
Title: RNA Sequencing of CRISPRi-TNFR1 Edited Human Nucleus Pulposus Cells Shows Shift of TNF-α from Pro-Inflammatory to Anti-Inflammatory via Potential TNFR2 Signaling
Abstract: Low back pain (LBP) is the leading cause of disability worldwide. Degeneration of the intervertebral disc (IVD), a condition known as degenerative disc disease (DDD), is among the prominent causes of LBP. DDD is characterized by a loss of disc height, a breakdown of the extracellular matrix, inflammation, and pain. Many treatment strategies for DDD and its associated pain have been proposed, but the underlying mechanisms remain largely unknown, making it challenging to develop therapeutics effectively targeting the degeneration and pain. Previously presented work demonstrated that lentiviral-delivered CRISPR epigenome editing of TNFR1 expression and TNF-α dosing in a rodent DDD model leads to protection against degeneration and disc height loss more robustly than TNFR1 editing alone. In this study, we have begun elucidating the underlying mechanisms of this work. Human nucleus pulposus (NP) cells were treated with TNFR1-editing vectors, with and without subsequent TNF-α dosing. RNA sequencing was performed and the top significantly altered metabolic pathways were determined. We demonstrated the myriad of gene expression changes that occur with TNFR1 editing and the changes of gene expression that occur with the addition of TNF-α dosing. The TNFR2 signaling pathway emerged as a top significantly altered metabolic pathway in both comparisons. Together, these results demonstrate that this additional protection against degeneration in the animal model could be due to a shift in signaling from TNFR1 to TNF-α’s other receptor, TNFR2, which has a predominantly anti-inflammatory effect.
Sushanto Kumar Saha
Co-authors: Nitish Khuran, Bretni Kennon, and William Niedermeyer
Faculty Advisor: Hamidreza Ghandehari
Title: An in vitro macrophage toxicity study of engineered high-energy silver nanoparticles
Abstract: EVQ-218 is a novel allotrope of silver nanoparticles developed via a patented nanosecond pulsed laser-based, high-pressure manufacturing process, that uses secondary laser systems to modify nanoparticles in the ejecta plume during particle formation. The surface properties of EVQ-218 show unique stability and antimicrobial properties without any ROS generation for cytotoxicity. Measuring up to the NIST standard, EVQ-218 has shown strong antibiofilm and antibacterial efficacy towards healthcare-associated infections (HAI) that can potentially benefit in mitigating medical device-related infections. These unique properties of EVQ-218 led to our current study to check the interaction of EVQ-218 with phagocytic cells like macrophages. In this study, EVQ-218 was evaluated for toxicity, uptake, and phagocytic activity against three different macrophage cell lines: RAW 264.7 murine macrophages, MH-S murine alveolar macrophages, and THP-1 human monocyte-like cells.
BMES 2025 will be held in October, 2025 in San Diego, CA.